Freeze Two
Freeze Two Release: February 2013
BAM files and other data
A cool DGRP paper
Epistasis dominates the genetic architecture of Drosophila quantitative traits
This paper compares the results of GWAS analysis using the DGRP, and extreme trait mapping using the flyland population—a population derived from 40 DGRP lines and bred to shuffle their alleles.
 |
|
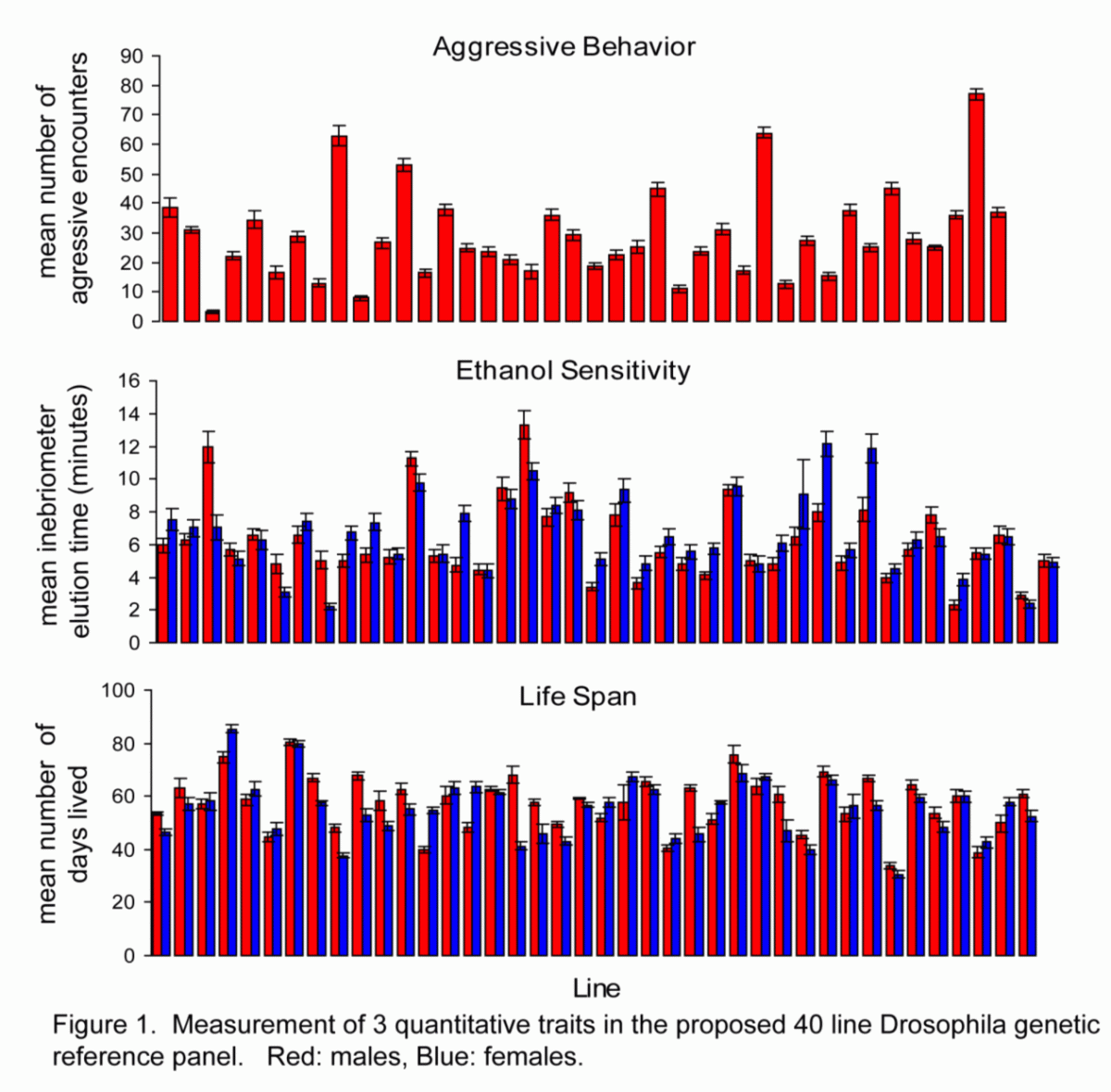
Measurements of Drosophila quantitative traits
Links
|
It finds that the overlap of putative quantitative trait alleles from the two methods is very small, but that we can detect extensive epistatic interaction between the alleles detected by both methods.
Networks of gene interactions can be detected which overlap with known gene networks and suggest new ones.
These networks probably represent the underlying genetic architecture of these traits better than a list of putative alleles.
Freeze One Publication
We are pleased to announce that freeze one of this great collaborative effort has been published in this paper:
The Drosophila melanogaster Genetic Reference Panel
Several companion papers also addressed this resource:
-
Joint genotyping on the fly: Identifying variation among a sequenced panel of inbred lines.
-
Using Whole-Genome Sequence Data to Predict Quantitative Trait Phenotypes in Drosophila melanogaster
We would like to thank everyone involved in this collaborative effort.
Freeze One Release: August 2010
We announced the freeze one release of the Drosophila Genetic Reference Panel under the Fort Lauderdale pre-publication data sharing agreement. We provided the Illumina sequence of 162 inbred D. melanogaster lines generated from a natural population in Raleigh North Carolina by the laboratory of Trudy Mackay.
Fastq Files
Each of these inbred lines has a minimum of 8.5X, but an average of 15X aligned genome sequence coverage in BAM files. Fastq files are available in the NCBI short read archive here.
DGRP Lines
All of the DGRP lines are available from the Bloomington Stock Center here. In the future we will be adding additional lines, and sequences, to increase the power of this genome-wide association toolkit.
For each line we also have individual links to the NCBI SRA, in this table, as in some cases multiple sequencing experiments are linked to the same DGRP line.
BAM Files
BCM-HGSC: Freeze One read alignments, stored as BAM files and generated with BWA
SNPs
For the Illumina reads we also provide a list of single nucleotide polymorphisms (SNPs) here generated by Eric Stone using a new algorithm that takes account of expected allele frequencies from 20 generations of full-sib inbreeding, followed by maintenance by random mating within each line.
Microsatellite Variability
This is a table of microsatellite variability across the 162 inbred, developed by David Mittelman.
Population Statistics
About DGRP
An amazing phenotypic resource: Our collaborator Trudy Mackay has a large set of > 200 inbred D. melanogaster lines with significant amounts of quantitative phenotypic data. These will form the basis of a community wide Drosophila Genetic Reference Panel (DGRP) where members of the community can perform phenotypic assays on a presequenced set of Drosophila lines for performing association studies at high power. This approach amortizes the sequencing costs over the entire Drosophila community. The figure provides data on three phenotypes, male aggressiveness, ethanol sensitivity and lifespan.
The Mackay laboratory also has data on life span, alcohol sensitivity, male aggression, male copulation latency, locomotive behavior, starvation resistance, chill coma recovery, abdominal bristle number, sternal pleural bristle number, adult mRNA transcription levels and olfactory behavior.
Additionally many other laboratories have promised to assay hundreds of additional phenotypes once sequence is available.
Community Involvement
Join our DGRP mailing list
Anyone can sign up for the public mailing list for DGRP-related discussion.
Data Release
As a service to the community we are releasing all sequence data for this project pre-publication and as soon as possible here—often pre-analysis on this ftp site. The data is released here under the standard genome sequence release agreements.
Users are free to use the data in scientific papers analyzing particular genes and regions if the providers of this data are properly acknowledged. Please cite the BCM-HGSC web site or publications from BCM-HGSC referring to the genome sequence. BCM-HGSC plans to publish the assembly and genomic annotation of the dataset, including genome wide molecular population genetic analyses, large-scale identification of regions of evolutionary conservation and quantitative trait association studies. This is in accordance with, and with the understandings in the Fort Lauderdale meeting discussing Community Resource Projects and the resulting NHGRI policy statement.
Data will also be deposited into the trace archive and other appropriate depositories once all QC/QA tests are passed.
DGRP line numbers correspond to Bloomington Drosophila Stock Center designations for these lines: e.g., DGRP_360 is the same as RAL-360.
Berkeley Drosophila Reference Sequencing Strain ((y[1]; cn[1] bw[1] sp[1]) b) Adams, M. D. et al. The genome sequence of Drosophila melanogaster. Science 287, 2185-95 (2000). This strain was kindly provided by Dr, Susan Celiniker at the BDGP. It is also available at the Bloomington Drosophila Stock Center.
DGRP line 375: 12X 454-long read (500bp) data, aligned sequence in fastm format. (The long read data was generated in house by 454 in Dec 2007 and Jan 2008 prior to the release of their next generation long read platform. We thank 454 Inc. for allowing us to release this data to the public, pre-publication.)